Method of adaptive knowledge distillation from multi-teacher to student deep learning models
Main Article Content
Abstract
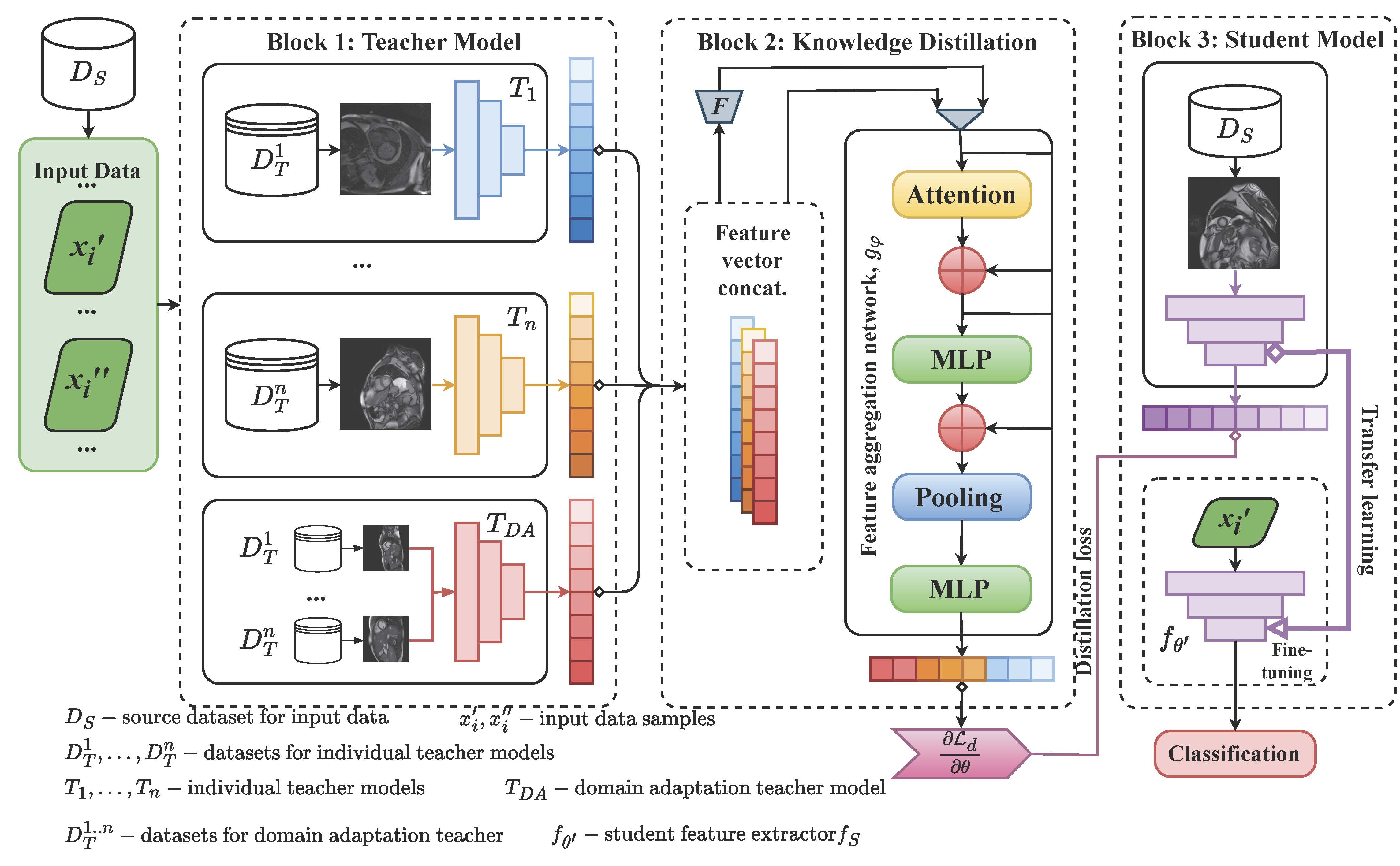
Transferring knowledge from multiple teacher models to a compact student model is often hindered by domain shifts between datasets and a scarcity of labeled target data, degrading performance. While existing methods address parts of this problem, a unified framework is lacking. In this work, we improve multi-teacher knowledge distillation by developing a holistic framework, enhanced multi-teacher knowledge distillation (EMTKD), that synergistically integrates three components: domain adaptation within teacher training, an instance-specific adaptive weighting mechanism for knowledge fusion, and semi-supervised learning to leverage unlabeled data. On a challenging cross-domain cardiac MRI benchmark, EMTKD achieves a target domain accuracy of 88.5% and an area under the curve of 92.5%, outperforming state-of-the-art techniques by up to 5.0%. Our results demonstrate that this integrated, adaptive approach yields significantly more robust and accurate student models, enabling effective deep learning deployment in data-scarce environments.
Downloads
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2025-07-10
Published 2025-11-21
References
Bernard, O., Lalande, A., Zotti, C., Cervenansky, F., Yang, X., Heng, P.A., Cetin, I., Lekadir, K., Camara, O., Gonzalez Ballester, M.A., Sanroma, G., Napel, S., Petersen, S., Tziritas, G., Grinias, E., Khened, M., Kollerathu, V.A., Krishnamurthi, G., Rohé, M.M., Pennec, X., Sermesant, M., Isensee, F., Jäger, P., Maier-Hein, K.H., Full, P.M., Wolf, I., Engelhardt, S., Baumgartner, C.F., Koch, L.M., Wolterink, J.M., Išgum, I., Jang, Y., Hong, Y., Patravali, J., Jain, S., Humbert, O. and Jodoin, P.M., 2018. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Transactions on Medical Imaging, 37(11), pp.2514–2525. Available from: https://doi.org/10.1109/TMI.2018.2837502. DOI: https://doi.org/10.1109/TMI.2018.2837502
Campello, V.M., Gkontra, P., Izquierdo, C., Martín-Isla, C., Sojoudi, A., Full, P.M., Maier-Hein, K., Zhang, Y., He, Z., Ma, J., Parreño, M., Albiol, A., Kong, F., Shadden, S.C., Acero, J.C., Sundaresan, V., Saber, M., Elattar, M., Li, H., Menze, B., Khader, F., Haarburger, C., Scannell, C.M., Veta, M., Carscadden, A., Punithakumar, K., Liu, X., Tsaftaris, S.A., Huang, X., Yang, X., Li, L., Zhuang, X., Viladés, D., Descalzo, M.L., Guala, A., Mura, L.L., Friedrich, M.G., Garg, R., Lebel, J., Henriques, F., Karakas, M., Çavu¸s, E., Petersen, S.E., Escalera, S., Seguí, S., Rodríguez-Palomares, J.F. and Lekadir, K., 2021. Multi-centre, multi-vendor and multi-disease cardiac segmentation: The M&Ms challenge. IEEE Transactions on Medical Imaging, 40(12), pp.3543–3554. Available from: https://doi.org/10.1109/TMI.2021.3090082. DOI: https://doi.org/10.1109/TMI.2021.3090082
Chaban, O., Manziuk, E., Markevych, O., Petrovskyi, S. and Radiuk, P., 2025. EMTKD at the edge: An adaptive multi-teacher knowledge distillation for robust cardiac MRI classification. In: T.A. Vakaliuk and S.O. Semerikov, eds. Proceedings of the 5th Edge Computing Workshop (doors 2025), Zhytomyr, Ukraine, April 4, 2025, CEUR Workshop Proceedings, vol. 3943. CEUR-WS.org, pp.42–57. Available from: https://ceur-ws.org/Vol-3943/paper09.pdf.
Chen, S., Bortsova, G., García-Uceda Juárez, A., Tulder, G. van der and Bruijne, M. de, 2019. Multi-task attention-based semi-supervised learning for medical image segmentation. In: D. Shen et al., eds. Medical Image Computing and Computer Assisted Intervention – MICCAI 2019, Lecture Notes in Computer Science, vol. 11766. Cham: Springer International Publishing, pp.457–465. Available from: https://doi.org/10.1007/978-3-030-32248-9_51. DOI: https://doi.org/10.1007/978-3-030-32248-9_51
Du, S., You, S., Li, X., Wu, J., Wang, F., Qian, C. and Zhang, C., 2020. Agree to disagree: Adaptive ensemble knowledge distillation in gradient space. Proceedings of the 34th International Conference on Neural Information Processing Systems. pp.12345–12355. Available from: https://dl.acm.org/doi/10.5555/3495724.3496759.
Ganin, Y. and Lempitsky, V.S., 2015. Unsupervised domain adaptation by backpropagation. In: F.R. Bach and D.M. Blei, eds. Proceedings of the 32nd International Conference on Machine Learning, JMLR Workshop and Conference Proceedings, vol. 37. JMLR.org, pp.1180–1189. Available from: https://dl.acm.org/doi/abs/10.5555/3045118.3045244.
Gao, H., Guo, J., Wang, G. and Zhang, Q., 2022. Cross-domain correlation distillation for unsupervised domain adaptation in nighttime semantic segmentation. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). pp.9903–9913. Available from: https://doi.org/10.1109/CVPR52688.2022.00968. DOI: https://doi.org/10.1109/CVPR52688.2022.00968
He, K., Zhang, X., Ren, S. and Sun, J., 2016. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). pp.770–778. Available from: https://doi.org/10.1109/cvpr.2016.90. DOI: https://doi.org/10.1109/CVPR.2016.90
Hesse, K., Khanji, M.Y., Aung, N., Dabbagh, G.S., Petersen, S.E. and Chahal, C.A.A., 2024. Assessing heterogeneity on cardiovascular magnetic resonance imaging: a novel approach to diagnosis and risk stratification in cardiac diseases. European Heart Journal – Cardiovascular Imaging, 25(4), pp.437–445. Available from: https://doi.org/10.1093/ehjci/jead285. DOI: https://doi.org/10.1093/ehjci/jead285
Hinton, G., Vinyals, O. and Dean, J., 2015. Distilling the knowledge in a neural network. 1503.02531, Available from: https://doi.org/10.48550/arXiv.1503.02531.
Kushol, R., Wilman, A.H., Kalra, S. and Yang, Y.H., 2023. DSMRI: Domain shift analyzer for multi-center MRI datasets. Diagnostics, 13(18), p.2947. Available from: https://doi.org/10.3390/diagnostics13182947. DOI: https://doi.org/10.3390/diagnostics13182947
Morales, M.A., Manning, W.J. and Nezafat, R., 2024. Present and future innovations in AI and cardiac MRI. Radiology, 310(1), p.e231269. Available from: https://doi.org/10.1148/radiol.231269. DOI: https://doi.org/10.1148/radiol.231269
Nabavi, S., Hamedani, K.A., Moghaddam, M.E., Abin, A.A. and Frangi, A.F., 2024. Multiple teachers-meticulous student: A domain adaptive meta-knowledge distillation model for medical image classification. 2403.11226, Available from: https://doi.org/10.48550/arXiv.2403.11226.
Nabavi, S., Hashemi, M., Moghaddam, M.E., Abin, A.A. and Frangi, A.F., 2024. Automated cardiac coverage assessment in cardiovascular magnetic resonance imaging using an explainable recurrent 3D dual-domain convolutional network. Medical Physics, 51(12), pp.8789–8803. Available from: https://doi.org/10.1002/mp.17411. DOI: https://doi.org/10.1002/mp.17411
Radiuk, P., Barmak, O.V., Manziuk, E.A. and Krak, I.V., 2024. Explainable Deep Learning: A Visual Analytics Approach with Transition Matrices. Mathematics, 12(7), p.1024. Available from: https://doi.org/10.3390/math12071024. DOI: https://doi.org/10.3390/math12071024
Rainio, O., Teuho, J. and Klén, R., 2024. Evaluation metrics and statistical tests for machine learning. Scientific Reports, 14(1), p.6086. Available from: https://doi.org/10.1038/s41598-024-56706-x. DOI: https://doi.org/10.1038/s41598-024-56706-x
Schmidhuber, J., 1992. Learning complex, extended sequences using the principle of history compression. Neural Computation, 4(2), pp.234–242. Available from: https://doi.org/10.1162/neco.1992.4.2.234. DOI: https://doi.org/10.1162/neco.1992.4.2.234
Shakor, M.Y. and Khaleel, M.I., 2025. Modern deep learning techniques for big medical data processing in cloud. IEEE Access, 13, pp.62005–62028. Available from: https://doi.org/10.1109/ACCESS.2025.3556327. DOI: https://doi.org/10.1109/ACCESS.2025.3556327
Singh, P. et al., 2022. One clinician is all you need–cardiac magnetic resonance imaging measurement extraction: Deep learning algorithm development. JMIR Medical Informatics, 10(9), p.e38178. Available from: https://doi.org/10.2196/38178. DOI: https://doi.org/10.2196/38178
Sohn, K., Berthelot, D., Carlini, N., Zhang, Z., Zhang, H., Raffel, C.A., Cubuk, E.D., Kurakin, A. and Li, C.L., 2020. FixMatch: Simplifying Semi-Supervised Learning with Consistency and Confidence. Proceedings of the 34th International Conference on Neural Information Processing Systems. pp.596–608. Available from: https://dl.acm.org/doi/abs/10.5555/3495724.3495775.
Tang, J., Chen, S., Niu, G., Sugiyama, M. and Gong, C., 2023. Distribution shift matters for knowledge distillation with webly collected images. 2023 IEEE/CVF International Conference on Computer Vision (ICCV). pp.17424–17434. Available from: https://doi.org/10.1109/ICCV51070.2023.01602. DOI: https://doi.org/10.1109/ICCV51070.2023.01602
Tang, J., Chen, S., Niu, G., Zhu, H., Zhou, J.T., Gong, C. and Sugiyama, M., 2024. Direct distillation between different domains. In: A. Leonardis, E. Ricci, S. Roth, O. Russakovsky, T. Sattler and G. Varol, eds. Computer Vision – ECCV 2024, Lecture Notes in Computer Science, vol. 15138. Cham: Springer Nature Switzerland, pp.154–172. Available from: https://doi.org/10.1007/978-3-031-72989-8_9. DOI: https://doi.org/10.1007/978-3-031-72989-8_9
Yang, C., Yu, X., Yang, H., An, Z., Yu, C., Huang, L. and Xu, Y., 2025. Multiteacher knowledge distillation with reinforcement learning for visual recognition. 2502.18510, Available from: https://doi.org/10.48550/arXiv.2502.18510.
Zhang, H., Chen, D. and Wang, C., 2022. Confidence-aware multi-teacher knowledge distillation. ICASSP 2022 - 2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). pp.4498–4502. Available from: https://doi.org/10.1109/ICASSP43922.2022.9747534. DOI: https://doi.org/10.1109/ICASSP43922.2022.9747534
Zhang, W., Zhu, L., Hallinan, J., Zhang, S., Makmur, A., Cai, Q. and Ooi, B.C., 2022. BoostMIS: Boosting medical image semi-supervised learning with adaptive pseudo labeling and informative active annotation. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, pp.20634–20644. Available from: https://doi.org/10.1109/CVPR52688.2022.02001. DOI: https://doi.org/10.1109/CVPR52688.2022.02001
Zhong, T., Chi, Z., Gu, L., Wang, Y., Yu, Y. and Tang, J., 2022. Meta-DMoE: Adapting to domain shift by meta-distillation from mixture-of-experts. Proceedings of the 36th International Conference on Neural Information Processing Systems. pp.22243–22257. Available from: https://dl.acm.org/doi/10.5555/3600270.3601886.